Vertica can import, export, and make predictions with PMML models of version 4.4 and below.
This is the multi-page printable view of this section. Click here to print.
Using PMML models
- 1: Exporting Vertica models in PMML format
- 2: Importing and predicting with PMML models
- 3: PMML features and attributes
1 - Exporting Vertica models in PMML format
You can take advantage of the built-in distributed algorithms in Vertica to train machine learning models. There might be cases in which you want to use these models for prediction outside Vertica, for example on an edge node. You can export certain Vertica models in PMML format and use them for prediction using a library or platform that supports reading and evaluating PMML models. Vertica supports the export of the following Vertica model types into PMML format: KMEANS, LINEAR_REGRESSION, LOGISTIC_REGRESSION, RF_CLASSIFIER, RF_REGRESSOR, XGB_CLASSIFIER, and XGB_REGRESSOR.
Here is an example for training a model in Vertica and then exporting it in PMML format. The following diagram shows the workflow of the example. We use vsql to run this example.
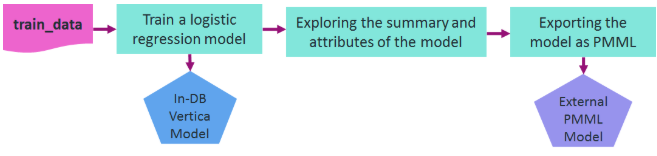
Let's assume that you want to train a logistic regression model on the data in a relation named 'patients' in order to predict the second attack of patients given their treatment and trait anxiety.
After training, the model is shown in a system table named V_CATALOG.MODELS which lists the archived ML models in Vertica.
=> -- Training a logistic regression model on a training_data
=> SELECT logistic_reg('myModel', 'patients', 'second_attack', 'treatment, trait_anxiety');
logistic_reg
---------------------------
Finished in 5 iterations
(1 row)
=> -- Looking at the models table
=> SELECT model_name, schema_name, category, model_type, create_time, size FROM models;
model_name | schema_name | category | model_type | create_time | size
------------+-------------+----------------+---------------------+-------------------------------+------
myModel | public | VERTICA_MODELS | LOGISTIC_REGRESSION | 2020-07-28 00:05:18.441958-04 | 1845
(1 row)
You can look at the summary of the model using the GET_MODEL_SUMMARY function.
=> -- Looking at the summary of the model
=> \t
Showing only tuples.
=> SELECT get_model_summary(USING PARAMETERS model_name='myModel');
=======
details
=======
predictor |coefficient|std_err |z_value |p_value
-------------+-----------+--------+--------+--------
Intercept | -6.36347 | 3.21390|-1.97998| 0.04771
treatment | -1.02411 | 1.17108|-0.87450| 0.38185
trait_anxiety| 0.11904 | 0.05498| 2.16527| 0.03037
==============
regularization
==============
type| lambda
----+--------
none| 1.00000
===========
call_string
===========
logistic_reg('public.myModel', 'patients', '"second_attack"', 'treatment, trait_anxiety'
USING PARAMETERS optimizer='newton', epsilon=1e-06, max_iterations=100, regularization='none', lambda=1, alpha=0.5)
===============
Additional Info
===============
Name |Value
------------------+-----
iteration_count | 5
rejected_row_count| 0
accepted_row_count| 20
You can also retrieve the model's attributes using the GET_MODEL_ATTRIBUTE function.
=> \t
Tuples only is off.
=> -- The list of the attributes of the model
=> SELECT get_model_attribute(USING PARAMETERS model_name='myModel');
attr_name | attr_fields | #_of_rows
--------------------+---------------------------------------------------+-----------
details | predictor, coefficient, std_err, z_value, p_value | 3
regularization | type, lambda | 1
iteration_count | iteration_count | 1
rejected_row_count | rejected_row_count | 1
accepted_row_count | accepted_row_count | 1
call_string | call_string | 1
(6 rows)
=> -- Returning the coefficients of the model in a tabular format
=> SELECT get_model_attribute(USING PARAMETERS model_name='myModel', attr_name='details');
predictor | coefficient | std_err | z_value | p_value
---------------+-------------------+--------------------+--------------------+--------------------
Intercept | -6.36346994178182 | 3.21390452471434 | -1.97998101463435 | 0.0477056620380991
treatment | -1.02410605239327 | 1.1710801464903 | -0.874496980810833 | 0.381847663704613
trait_anxiety | 0.119044916668605 | 0.0549791755747139 | 2.16527285875412 | 0.0303667955962211
(3 rows)
You can use the EXPORT_MODELS function in a simple statement to export the model in PMML format, as shown below.
=> -- Exporting the model as PMML
=> SELECT export_models('/data/username/temp', 'myModel' USING PARAMETERS category='PMML');
export_models
---------------
Success
(1 row)
See also
2 - Importing and predicting with PMML models
As a Vertica user, you can train ML models in other platforms, convert them to standard PMML format, and then import them into Vertica for in-database prediction on data stored in Vertica relations.
Here is an example of how to import a PMML model trained in Spark. The following diagram shows the workflow of the example.

You can use the IMPORT_MODELS function in a simple statement to import the PMML model. The imported model then appears in a system table named V_CATALOG.MODELS which lists the archived ML models in Vertica.
=> -- importing the PMML model trained and generated in Spark
=> SELECT import_models('/data/username/temp/spark_logistic_reg' USING PARAMETERS category='PMML');
import_models
---------------
Success
(1 row)
=> -- Looking at the models table=> SELECT model_name, schema_name, category, model_type, create_time, size FROM models;
model_name | schema_name | category | model_type | create_time | size
--------------------+-------------+----------+-----------------------+-------------------------------+------
spark_logistic_reg | public | PMML | PMML_REGRESSION_MODEL | 2020-07-28 00:12:29.389709-04 | 5831
(1 row)
You can look at the summary of the model using GET_MODEL_SUMMARY function.
=> \t
Showing only tuples.
=> SELECT get_model_summary(USING PARAMETERS model_name='spark_logistic_reg');
=============
function_name
=============
classification
===========
data_fields
===========
name |dataType| optype
-------+--------+-----------
field_0| double |continuous
field_1| double |continuous
field_2| double |continuous
field_3| double |continuous
field_4| double |continuous
field_5| double |continuous
field_6| double |continuous
field_7| double |continuous
field_8| double |continuous
target | string |categorical
==========
predictors
==========
name |exponent|coefficient
-------+--------+-----------
field_0| 1 | -0.23318
field_1| 1 | 0.73623
field_2| 1 | 0.29964
field_3| 1 | 0.12809
field_4| 1 | -0.66857
field_5| 1 | 0.51675
field_6| 1 | -0.41026
field_7| 1 | 0.30829
field_8| 1 | -0.17788
===============
Additional Info
===============
Name | Value
-------------+--------
is_supervised| 1
intercept |-1.20173
You can also retrieve the model's attributes using the GET_MODEL_ATTRIBUTE function.
=> \t
Tuples only is off.
=> -- The list of the attributes of the PMML model
=> SELECT get_model_attribute(USING PARAMETERS model_name='spark_logistic_reg');
attr_name | attr_fields | #_of_rows
---------------+-----------------------------+-----------
is_supervised | is_supervised | 1
function_name | function_name | 1
data_fields | name, dataType, optype | 10
intercept | intercept | 1
predictors | name, exponent, coefficient | 9
(5 rows)
=> -- The coefficients of the PMML model
=> SELECT get_model_attribute(USING PARAMETERS model_name='spark_logistic_reg', attr_name='predictors');
name | exponent | coefficient
---------+----------+--------------------
field_0 | 1 | -0.2331769167607
field_1 | 1 | 0.736227459496199
field_2 | 1 | 0.29963728232024
field_3 | 1 | 0.128085369856188
field_4 | 1 | -0.668573096260048
field_5 | 1 | 0.516750679584637
field_6 | 1 | -0.41025989394959
field_7 | 1 | 0.308289533913736
field_8 | 1 | -0.177878773139411
(9 rows)
You can then use the PREDICT_PMML function to apply the imported model on a relation for in-database prediction. The internal parameters of the model can be matched to the column names of the input relation by their names or their listed position. Direct input values can also be fed to the function as displayed below.
=> -- Using the imported PMML model for scoring direct input values
=> SELECT predict_pmml(1.5,0.5,2,1,0.75,4.2,3.1,0.9,1.1
username(> USING PARAMETERS model_name='spark_logistic_reg', match_by_pos=true);
predict_pmml
--------------
1
(1 row)
=> -- Using the imported PMML model for scoring samples in a table
=> SELECT predict_pmml(* USING PARAMETERS model_name='spark_logistic_reg') AS prediction
=> FROM test_data;
prediction
------------
1
0
(2 rows)
See also
3 - PMML features and attributes
To be compatible with Vertica, PMML models must conform to the following:
-
The PMML model does not have a data preprocessing step.
-
The encoded PMML model type is ClusteringModel, GeneralRegressionModel, MiningModel, RegressionModel, or TreeModel.
Supported PMML tags and attributes
The following table lists supported PMML tags and attributes.
| XML-tag name | Ignored attributes | Supported attributes | Unsupported attributes | Ignored sub-tags | Supported sub-tags | Unsupported sub-tags |
|---|---|---|---|---|---|---|
| Categories | - | - | - | - | Category | Extension |
| Category | - | value (required) | - | - | - | Extension |
| CategoricalPredictor | - |
|
- | - | - | Extension |
| Cluster | size |
|
- |
|
NUM-ARRAY |
|
| ClusteringField | - |
|
|
- | - |
|
| ClusteringModel | modelName |
|
- | ModelVerification |
|
|
| ComparisonMeasure |
|
|
- | - |
|
|
| CompoundPredicate | - | booleanOperator (required) | - | - | Extension | |
| CovariateList | - | - | - | - | Predictor | Extension |
| DataDictionary | - | numberOfFields | - | - | DataField |
|
| DataField | displayName |
|
|
- | Value |
|
| DerivedField | displayName |
|
- | - | FieldRef | |
| FactorList | - | - | - | - | Predictor | Extension |
| False | - | - | - | - | - | Extension |
| FieldRef | - | name (required) | mapMissingTo | - | - | Extension |
| GeneralRegressionModel |
|
|
|
|
|
|
| Header |
|
- | - |
|
- | - |
| LocalTransformations | - | - | - | - | DerivedField | Extension |
| MiningField |
|
|
|
- | - | Extension |
| MiningModel |
|
|
- | ModelVerification | ||
| MiningSchema | - | - | - | - | MiningField | Extension |
| Node | - |
|
- | - | ||
| NumericPredictor | - |
|
- | - | - | Extension |
| Output | - | - | - | - | OutputField | Extension |
| OutputField |
|
|
|
- | - |
|
| Parameter | - |
|
referencePoint | - | - | Extension |
| ParameterList | - | - | - | - | Parameter | Extension |
| ParamMatrix | - | - | - | - | PCell | Extension |
| PCell | - |
|
- | - | - | Extension |
| PPCell | - |
|
- | - | - | Extension |
| PPMatrix | - | - | - | - | PPCell | Extension |
| PMML | - |
|
- | MiningBuildTask |
|
|
| Predictor | - |
|
- | - | Extension | |
| RegressionModel |
|
|
- | ModelVerification |
|
|
| RegressionTable | - |
|
- | - |
|
|
| Segment | - |
|
- | - | ||
| Segmentation | - |
|
- | - | Segment | Extension |
| SimplePredicate | - |
|
- | - | - | Extension |
| SimpleSetPredicate | - |
|
- | - | ARRAY | Extension |
| Target | - |
|
|
- | - | |
| Targets | - | - | - | - | Target | Extension |
| TreeModel |
|
|
- | ModelVerification | ||
| True | - | - | - | - | - | Extension |
| Value | displayValue |
|
- | - | - | Extension |